Preparing and Investigating Data
Overview
Teaching: 60 min
Exercises: 30 minQuestions
What to do before you can bring the data into R?
Objectives
Identify different data types
Recognise different types of visualisations
First, we need to load the tidyverse package (actually a collection of packages).
library(tidyverse)
We can now load the Pattani data from the Excel file. For the notes, this is stored
in the data directory. It might be different for you.
path <- file.path("..", "data", "Pattani.xlsx")
pattani <- readxl::read_excel(path, sheet = "Data", skip = 1)
readxl::read_excel is used to explicitly call the read_excel function in the readxl
package.
We would like to see what the data looks like now that it has been read into R. You can just type the name of the dataset.
pattani
# A tibble: 434 x 8
ID `blood lead` age gender school duration water `ln(blood lead)`
<dbl> <dbl> <dbl> <chr> <chr> <dbl> <chr> <dbl>
1 1 11.7 13 Boy Tangkad~ 13 Boil 2.46
2 2 11.8 13 Boy Tangkad~ 5 Boil 2.47
3 3 6.4 13 Girl Tangkad~ 13 Stand 1.86
4 4 6.9 11 Girl Tangkad~ 11 Boil 1.93
5 5 10.3 13 Girl Tangkad~ 5 Boil 2.33
6 6 8.3 13 Girl Tangkad~ 13 Filt~ 2.12
7 7 6.2 13 Girl Tangkad~ 5 Filt~ 1.82
8 8 9.4 13 Boy Tangkad~ 13 Stand 2.24
9 9 14.9 11 Boy Tangkad~ 11 Boil 2.70
10 10 8.1 11 Girl Tangkad~ 11 Filt~ 2.09
# ... with 424 more rows
Read in the cuckoo data
Use the
read_csvfunction in thereadrpackage to loadcuckoo.csv.Solution
cuckoo <- readr::read_csv(file.path("..", "data", "cuckoo.csv")) cuckoo# A tibble: 45 x 2 Nest Length <chr> <dbl> 1 Robin 21.8 2 Robin 23.9 3 Robin 23 4 Robin 22.3 5 Robin 23.3 6 Robin 22 7 Robin 22.4 8 Robin 22.6 9 Robin 22.4 10 Robin 22 # ... with 35 more rows
Having names with spaces in them can make them more difficult to work with. We can add to the way that we read the file to modify the names.
pattani <- readxl::read_excel(path, sheet = "Data", skip = 1) %>%
rename(blood_lead = `blood lead`, ln_blood_lead = `ln(blood lead)`)
pattani
# A tibble: 434 x 8
ID blood_lead age gender school duration water ln_blood_lead
<dbl> <dbl> <dbl> <chr> <chr> <dbl> <chr> <dbl>
1 1 11.7 13 Boy Tangkadeng 13 Boil 2.46
2 2 11.8 13 Boy Tangkadeng 5 Boil 2.47
3 3 6.4 13 Girl Tangkadeng 13 Stand 1.86
4 4 6.9 11 Girl Tangkadeng 11 Boil 1.93
5 5 10.3 13 Girl Tangkadeng 5 Boil 2.33
6 6 8.3 13 Girl Tangkadeng 13 Filter 2.12
7 7 6.2 13 Girl Tangkadeng 5 Filter 1.82
8 8 9.4 13 Boy Tangkadeng 13 Stand 2.24
9 9 14.9 11 Boy Tangkadeng 11 Boil 2.70
10 10 8.1 11 Girl Tangkadeng 11 Filter 2.09
# ... with 424 more rows
What does
%>%mean?
%>%is something from thetidyversecalled a pipe. It allows you to take the output from 1 function (in this caseread_excel) and put it directly into another function (rename). It is as though there is a pipe connecting the functions together.
Simple summary statistics
Previously we got a preview of the data, but we might want to get some summary statistics
about the columns. You can use the function summary for this.
summary(pattani)
ID blood_lead age gender
Min. : 1.0 Min. : 4.00 Min. : 5.000 Length:434
1st Qu.:116.2 1st Qu.: 8.70 1st Qu.: 8.000 Class :character
Median :231.5 Median :11.70 Median : 9.000 Mode :character
Mean :229.5 Mean :12.05 Mean : 9.281
3rd Qu.:344.8 3rd Qu.:14.70 3rd Qu.:11.000
Max. :454.0 Max. :28.30 Max. :13.000
NA's :1
school duration water ln_blood_lead
Length:434 Min. : 2.000 Length:434 Min. :1.386
Class :character 1st Qu.: 5.000 Class :character 1st Qu.:2.163
Mode :character Median : 8.000 Mode :character Median :2.460
Mean : 7.922 Mean :2.425
3rd Qu.:10.000 3rd Qu.:2.688
Max. :13.000 Max. :3.343
NA's :1
Have a look at the character columns. What sort of variables are these?
If we convert these to factor then R knows that they are categorical variables and does
some helpful things for us. We will use the as_factor function, which is part of the
tidyverse. There is also a base R function as.factor (note the . instead of _)
but we will use the tidyverse version.
pattani <- readxl::read_excel(path, sheet = "Data", skip = 1) %>%
rename(blood_lead = `blood lead`, ln_blood_lead = `ln(blood lead)`) %>%
mutate(gender = as_factor(gender), school = as_factor(school), water = as_factor(water))
What happens if we run the summary now?
summary(pattani)
ID blood_lead age gender
Min. : 1.0 Min. : 4.00 Min. : 5.000 Boy :204
1st Qu.:116.2 1st Qu.: 8.70 1st Qu.: 8.000 Girl:230
Median :231.5 Median :11.70 Median : 9.000
Mean :229.5 Mean :12.05 Mean : 9.281
3rd Qu.:344.8 3rd Qu.:14.70 3rd Qu.:11.000
Max. :454.0 Max. :28.30 Max. :13.000
NA's :1
school duration water ln_blood_lead
Tangkadeng:136 Min. : 2.000 Boil :163 Min. :1.386
Thamthalu : 46 1st Qu.: 5.000 Stand :163 1st Qu.:2.163
Tachi :128 Median : 8.000 Filter : 48 Median :2.460
Tesabal 3 : 62 Mean : 7.922 Nothing: 56 Mean :2.425
Sabarang : 62 3rd Qu.:10.000 NA's : 4 3rd Qu.:2.688
Max. :13.000 Max. :3.343
NA's :1
The factor variables now show the number of each element in the variables.
Convert cuckoo data to a factor
Convert the
Nestcolumn to a factor and display the summary.Solution
cuckoo <- readr::read_csv(file.path("..", "data", "cuckoo.csv")) %>% mutate(Nest = as_factor(Nest)) summary(cuckoo)Nest Length Robin :16 Min. :19.80 Wren :15 1st Qu.:21.20 Sparrow:14 Median :22.30 Mean :22.24 3rd Qu.:23.00 Max. :25.00
Visualising the data
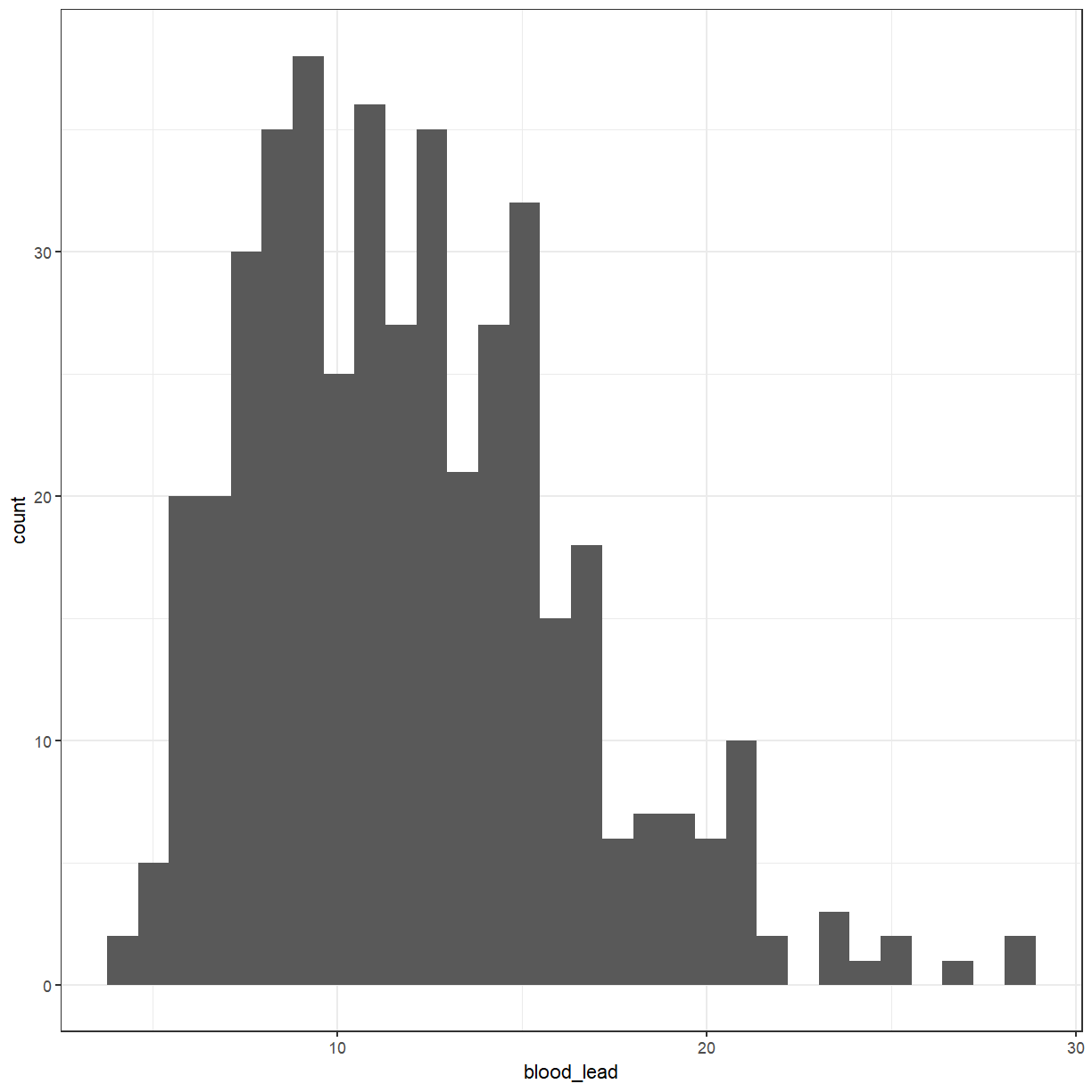
We will start with a histogram of the levels of lead in the blood.
ggplot(pattani, aes(x = blood_lead)) +
geom_histogram()
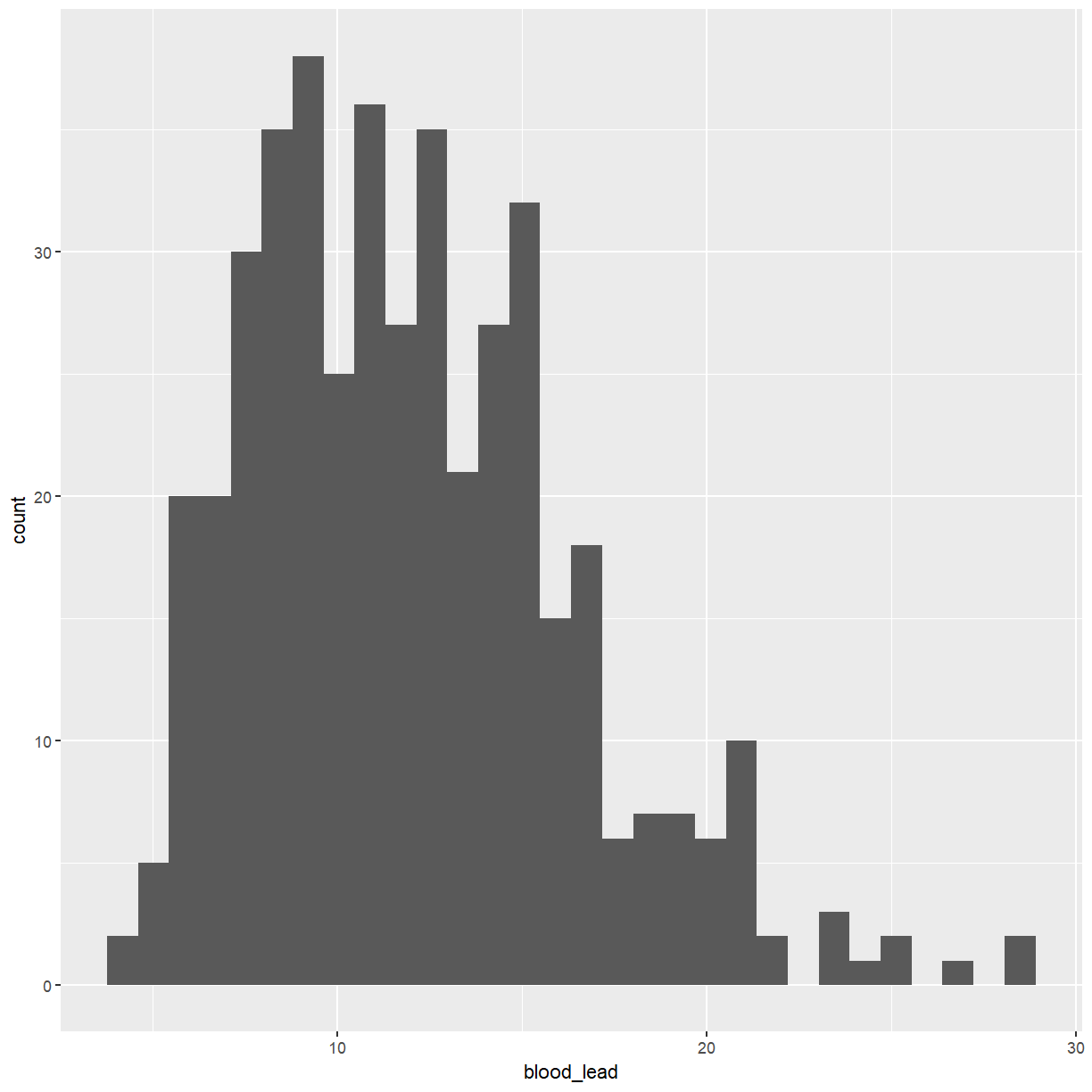
I am not a fan of the grey background that is the default with ggplot graphs. You can
change that by adding a theme.
ggplot(pattani, aes(x = blood_lead)) +
geom_histogram() +
theme_bw()
I will set the default theme so that I don’t have to specify it each time.
theme_set(theme_bw())
Create a histogram of the cuckoo egg lengths
Create a histogram of the cuckoo egg lengths
Solution
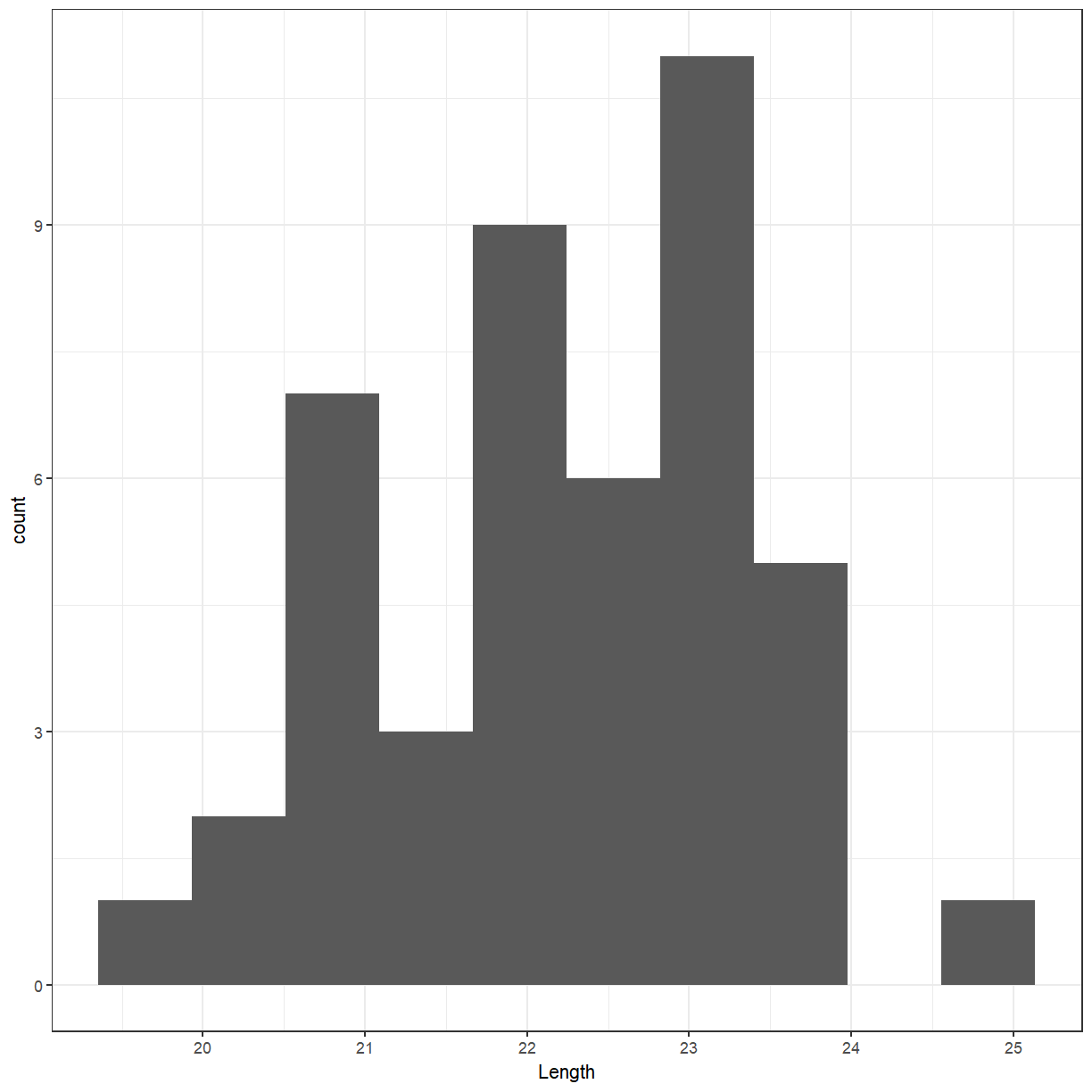
ggplot(cuckoo, aes(x = Length)) + geom_histogram(bins = 10)
Distribution?
- The distribution of a variable tells us what values it takes and how often it takes these values.
- When examining a distribution, look for shape, center, and spread and for clear deviations from the overall shape.
Moore, McCabe & Craig (2017)
You have probably heard of the Normal distribution, which has a bell shape.
ggplot(data = data.frame(x = c(-3, 3)), aes(x)) +
stat_function(fun = dnorm, n = 101, args = list(mean = 0, sd = 1)) + ylab("") +
scale_y_continuous(breaks = NULL)
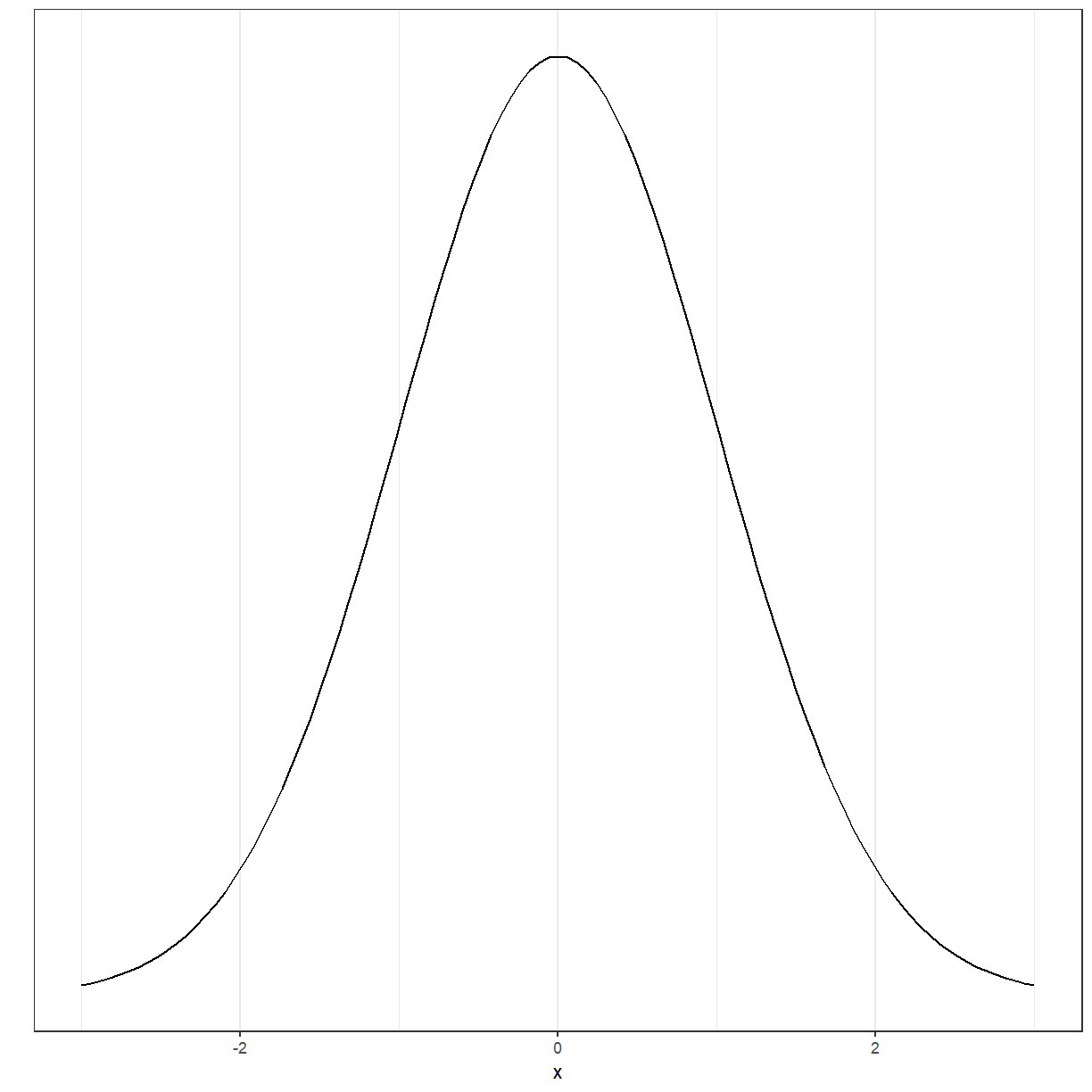
There are several other distributions. Several of the tests and models that we will discuss in this workshop have as one of the assumptions that either the data or the residuals of the model are Normally distributed.
Now back to what we were doing
For a lot of the statistical tests that we will be doing later, we would like the variable to be normally distributed. Looking at the histogram, it should be symmetric and bell-shaped.
ggplot(pattani, aes(x = ln_blood_lead)) +
geom_histogram()
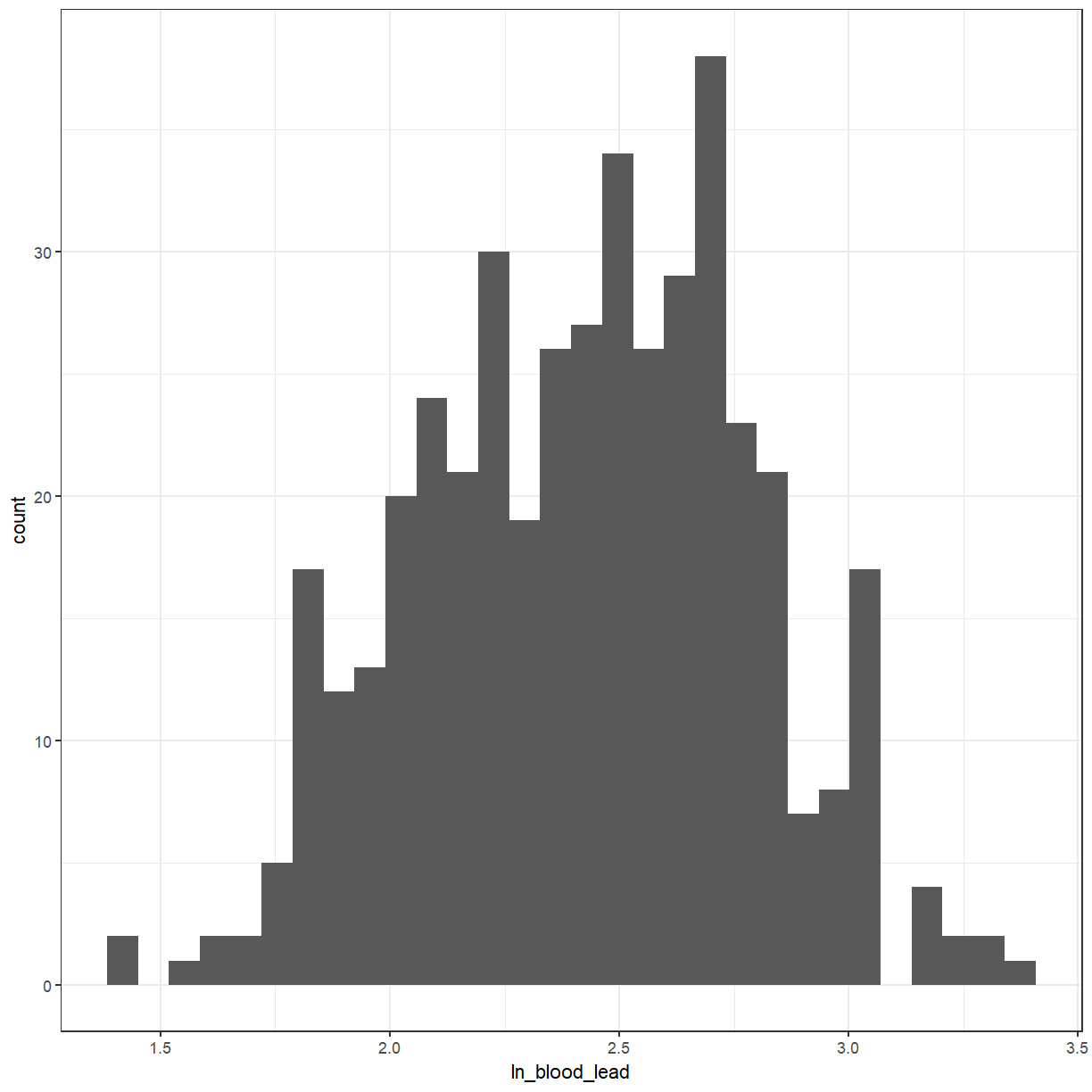
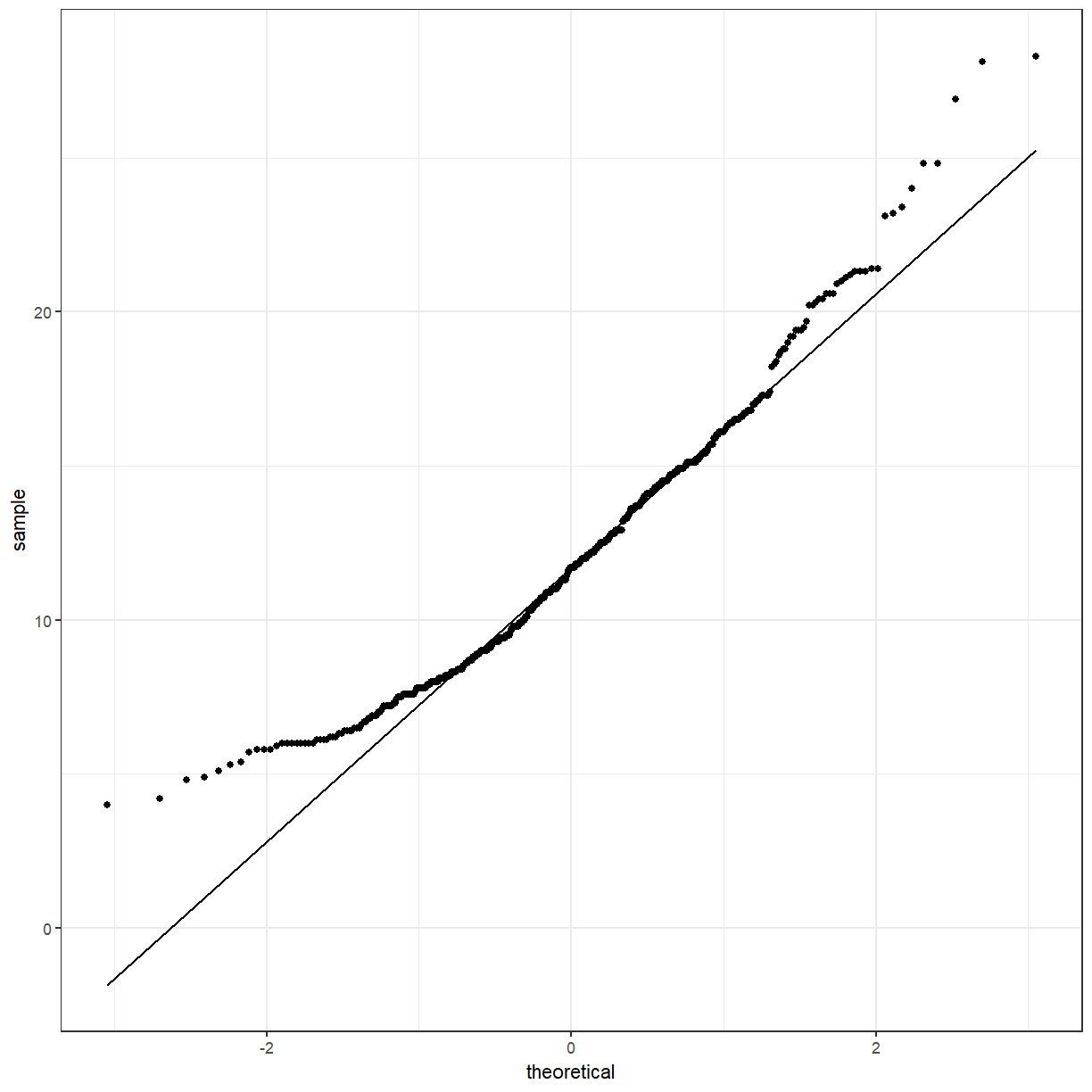
It looks a little bit better. We can also use a QQ-plot to check for normality. The points should lie approximately along the line.
ggplot(pattani, aes(sample = blood_lead)) +
geom_qq() +
geom_qq_line()
ggplot(pattani, aes(sample = ln_blood_lead)) +
geom_qq() +
geom_qq_line()
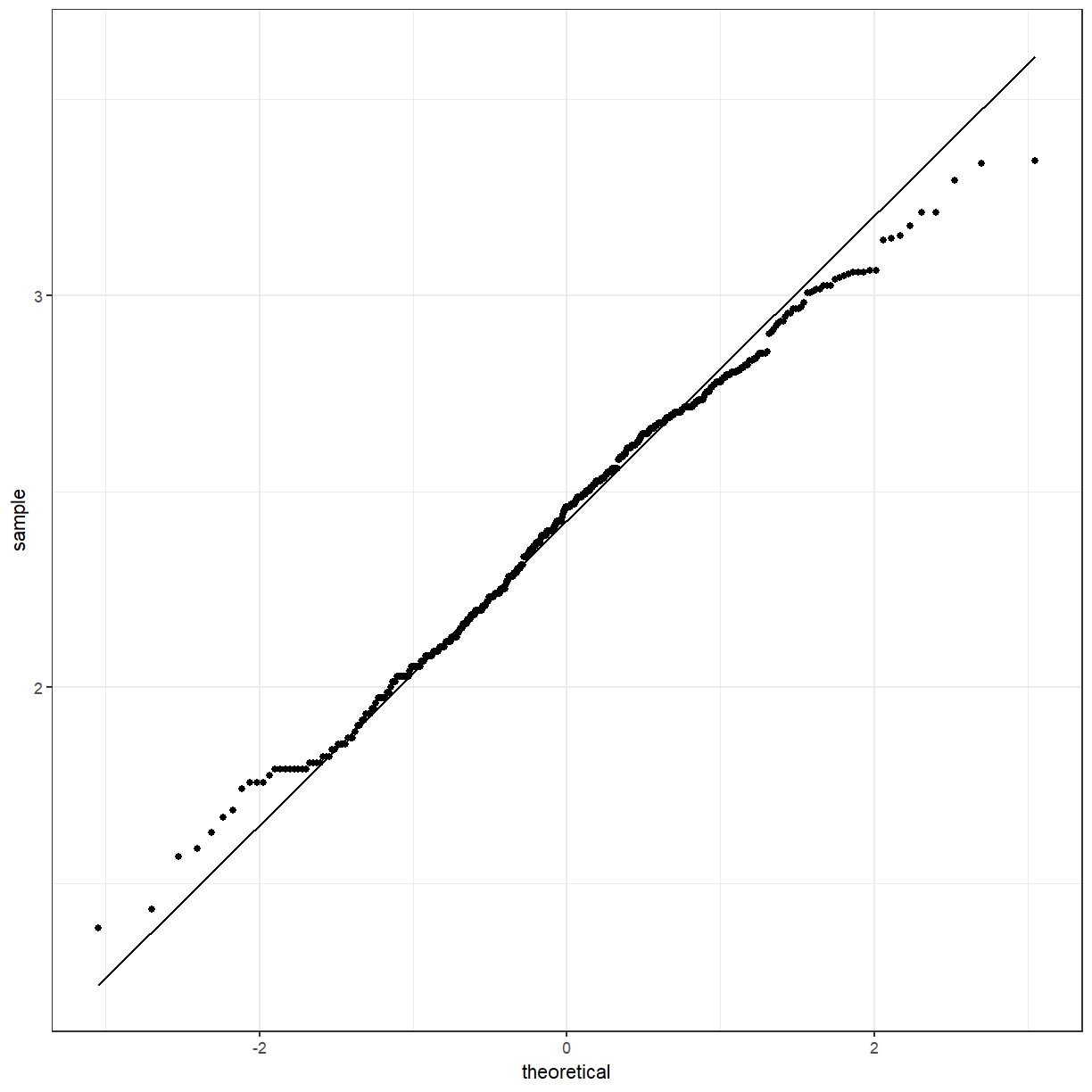
What if we wanted to see the difference between the boys and girls?
ggplot(pattani, aes(x = blood_lead, fill = gender)) +
geom_histogram()
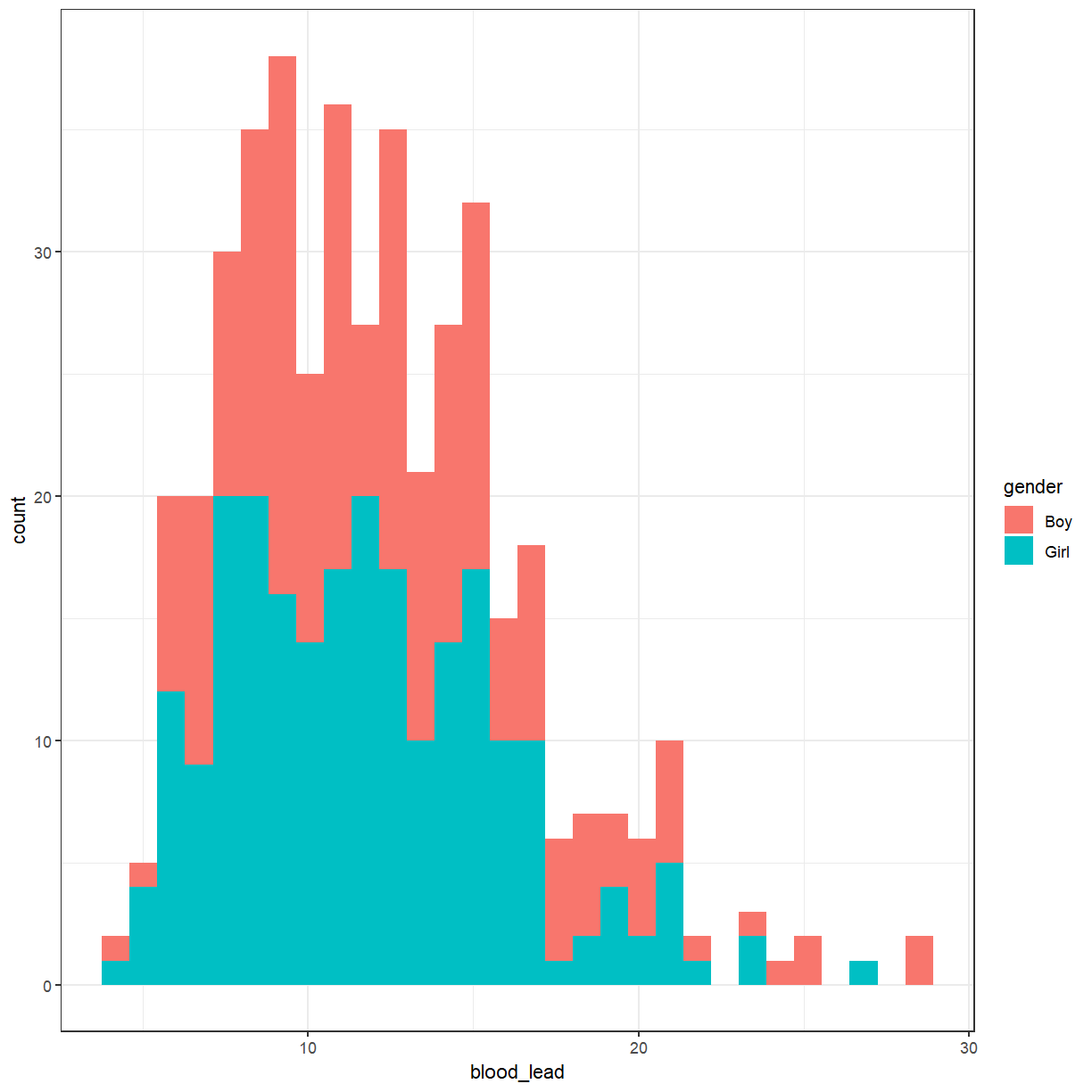
That produced a stacked histogram, which isn’t all that easy to read. It might be better if they were side-by-side.
ggplot(pattani, aes(x = blood_lead, fill = gender)) +
geom_histogram(position = "dodge")
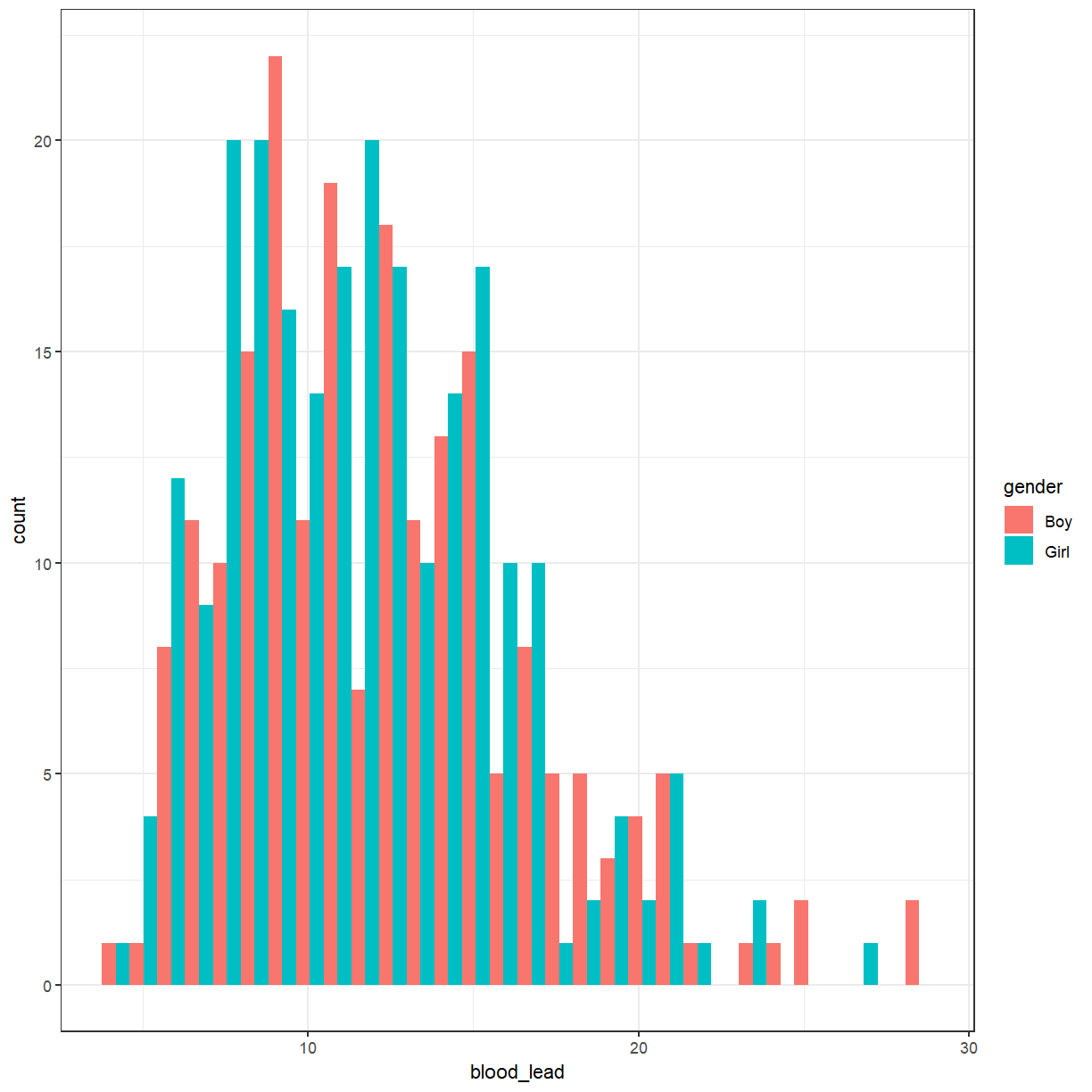
Cuckoo egg lengths by host nest
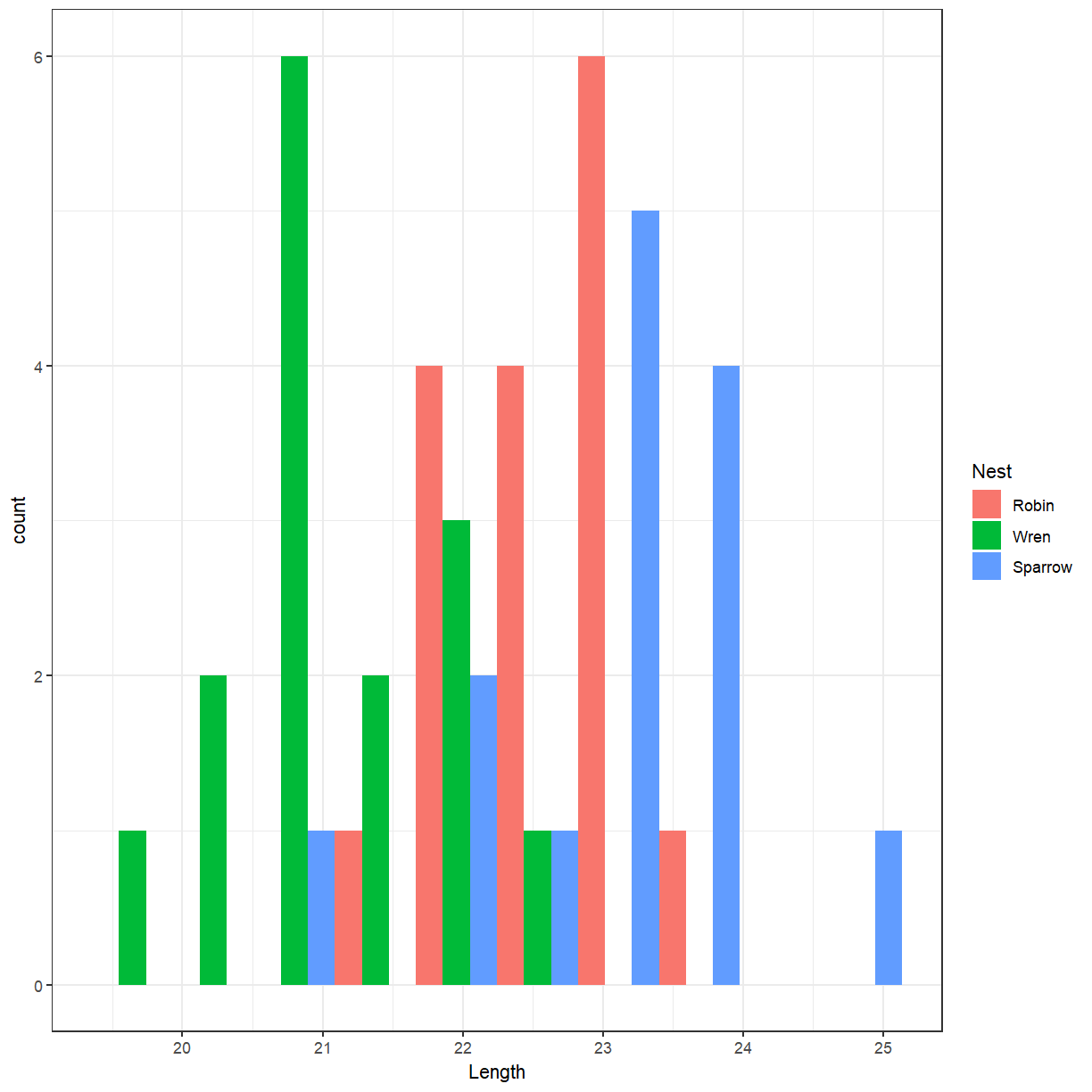
Create a histogram of the cuckoo egg lengths with colours showing the host nest species.
Solution
ggplot(cuckoo, aes(x = Length, fill = Nest)) + geom_histogram(position = "dodge", bins = 10)
There doesn’t seem to be much difference. Perhaps we should try a box plot. Notice that
gender is now the x variable and blood_lead is the y variable.
ggplot(pattani, aes(x = gender, y = blood_lead)) +
geom_boxplot()
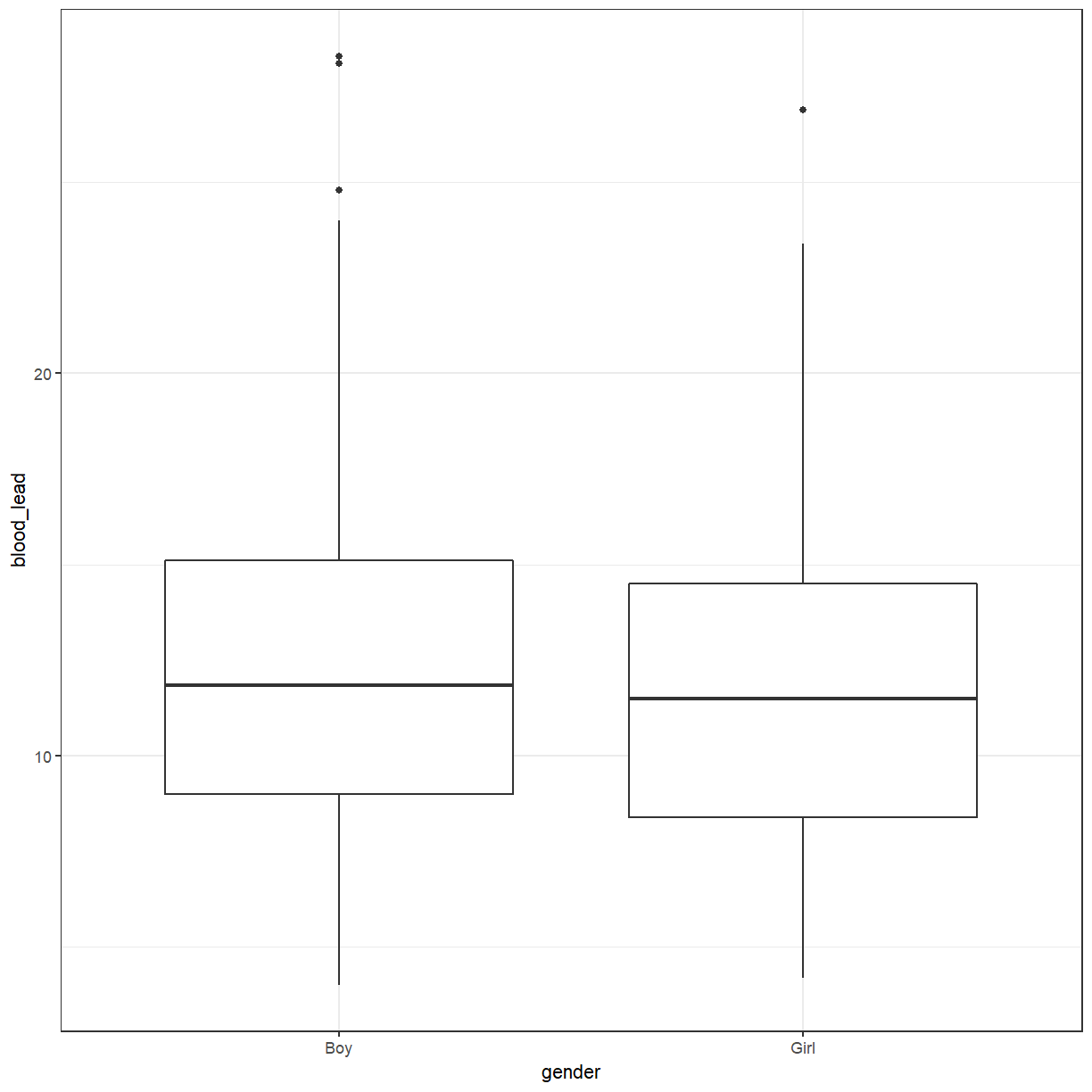
The box plot also suggests that there isn’t much difference between boys and girls. What about between schools?
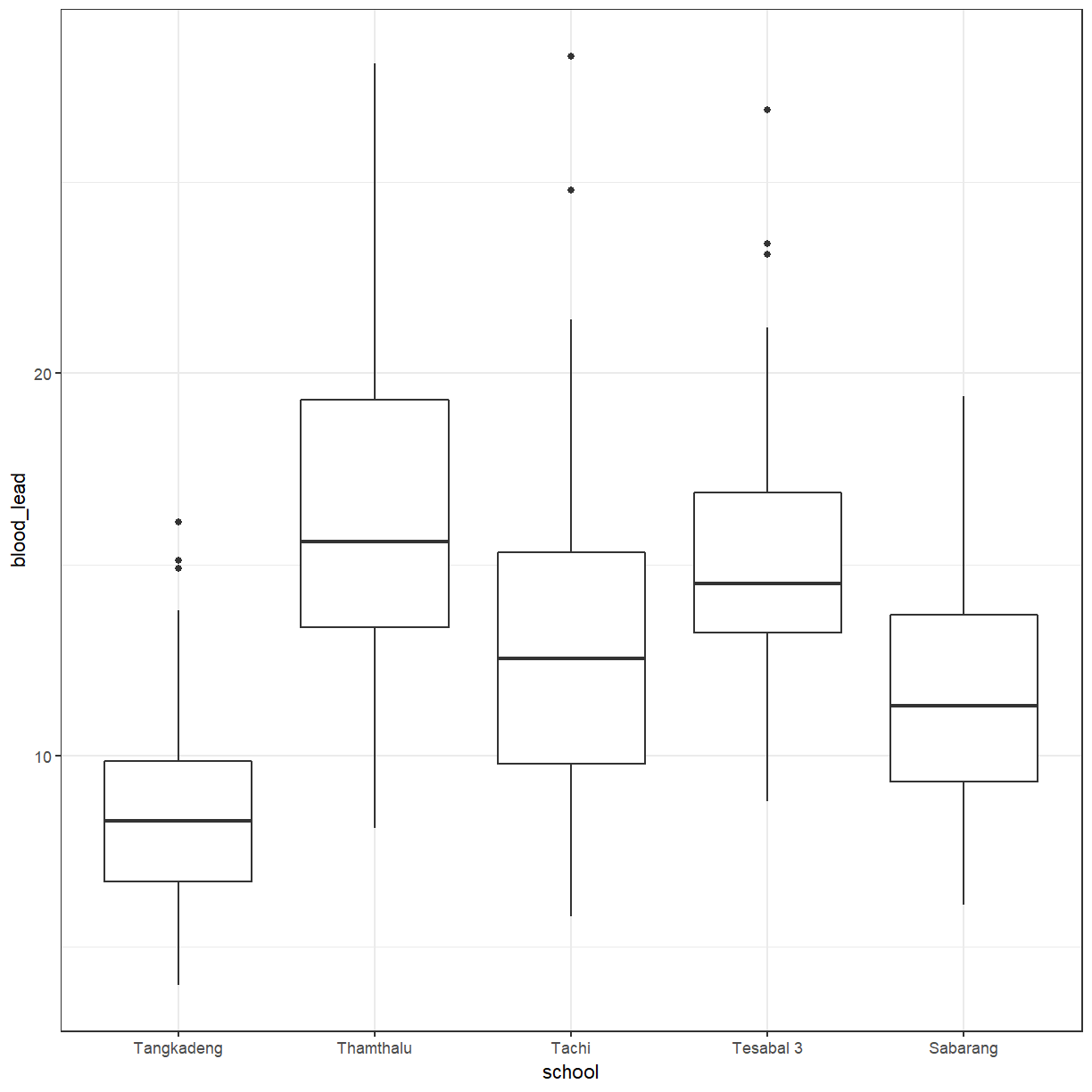
ggplot(pattani, aes(x = school, y = blood_lead)) +
geom_boxplot()
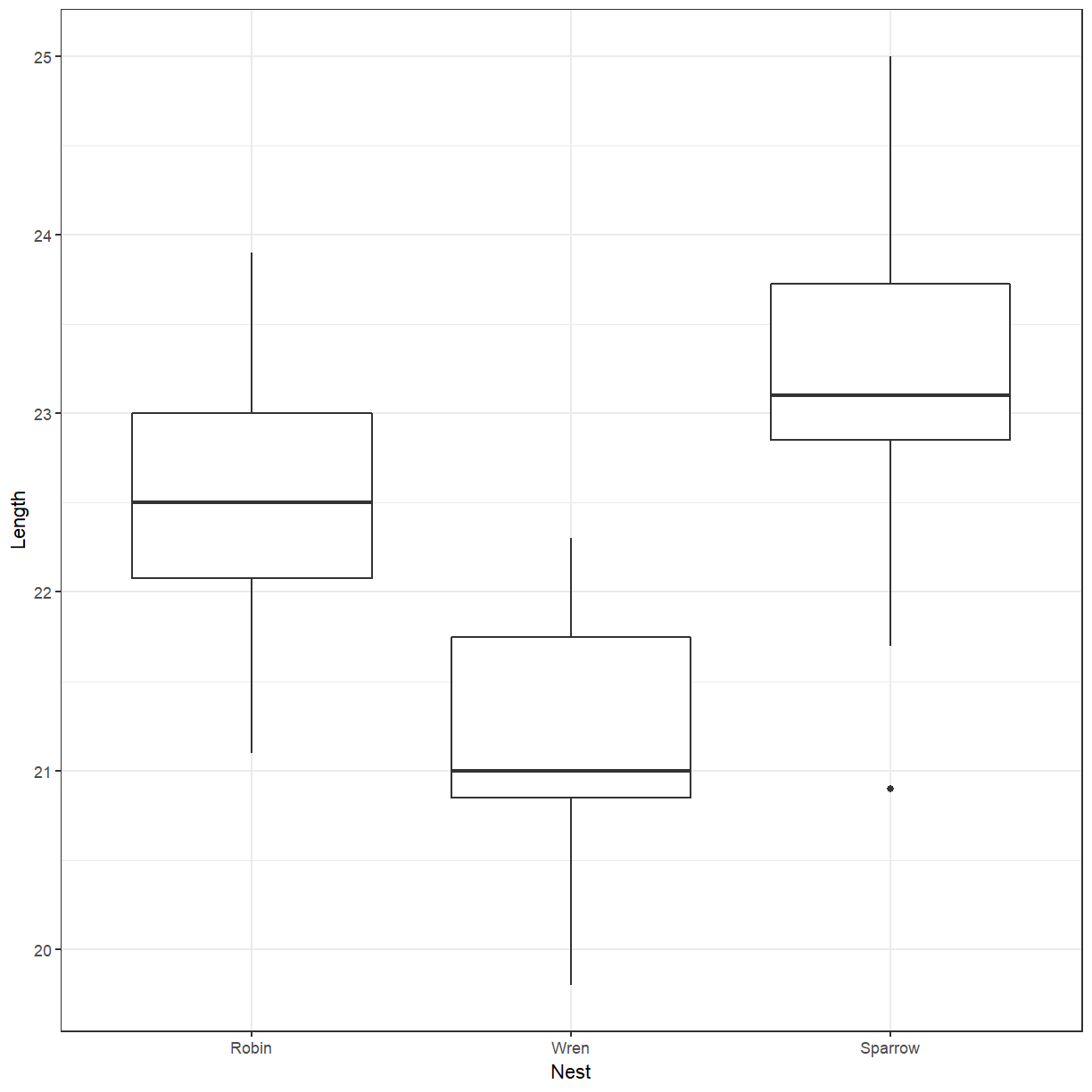
Box plot of cuckoo egg lengths by host nest
Create a box plot of the cuckoo egg lengths with colours showing the host nest species.
Solution
ggplot(cuckoo, aes(x = Nest, y = Length)) + geom_boxplot()
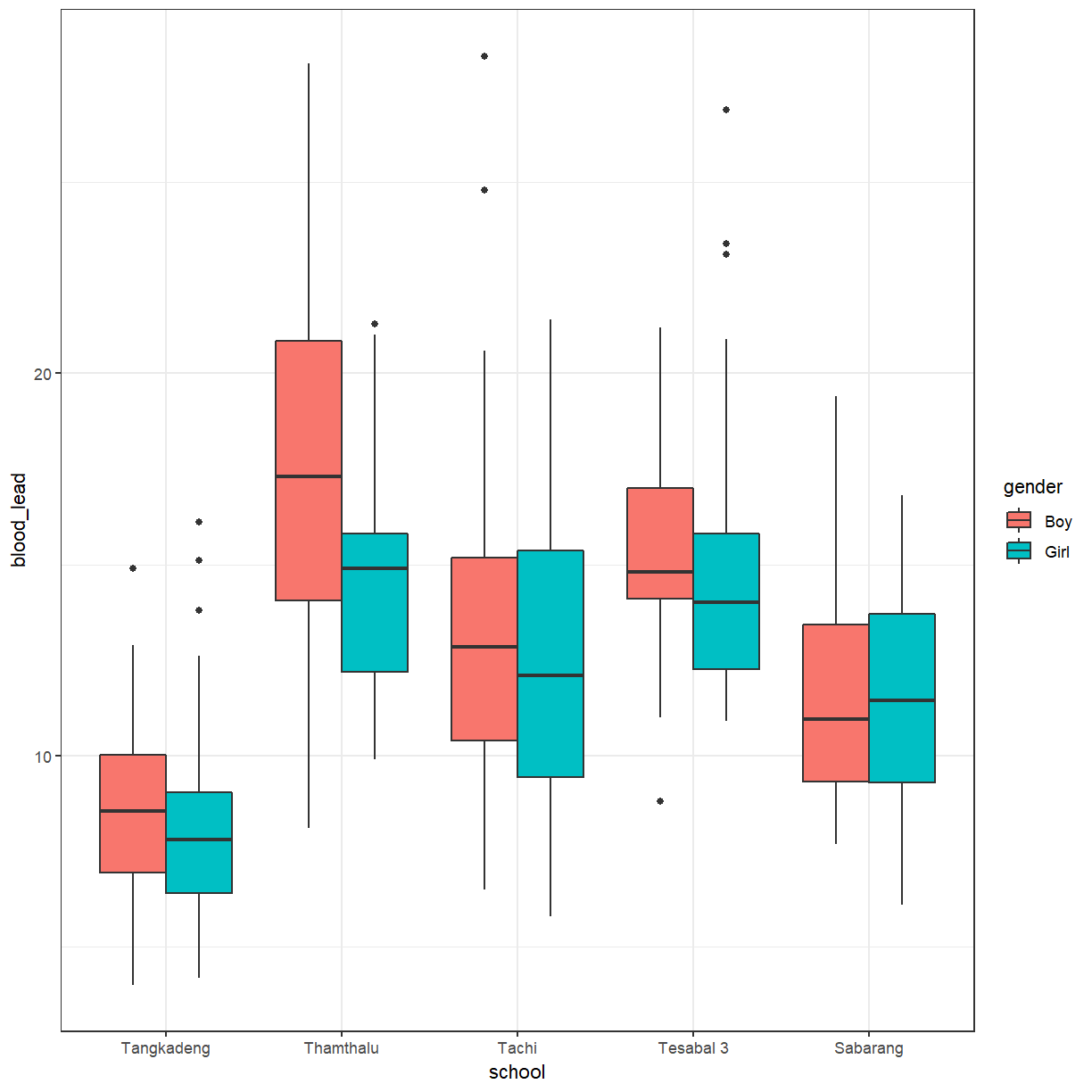
There does seems to be more difference between schools than between genders. We can add
gender back in again. Again, we have to use position = "dodge".
ggplot(pattani, aes(x = school, y = blood_lead, fill = gender)) +
geom_boxplot(position = "dodge")
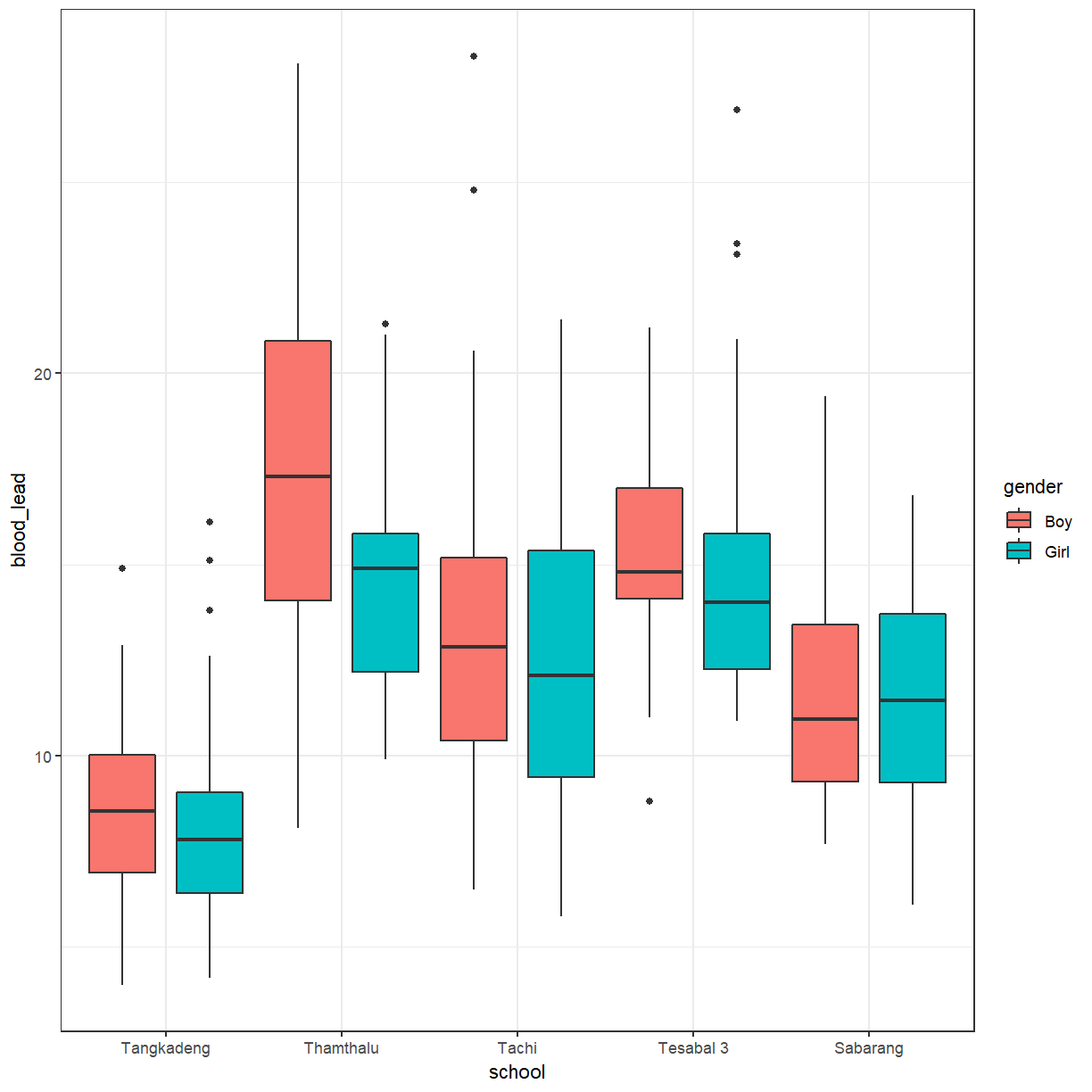
You can add a little bit of space between the boxes.
ggplot(pattani, aes(x = school, y = blood_lead, fill = gender)) +
geom_boxplot(position = position_dodge(1))
Key Points
Data should have 1 observation per row.